ATP-binding site of CDKs
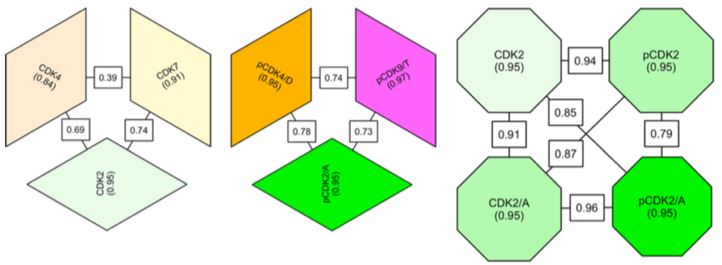
A significant requirement for the further development of therapeutically useful compounds is a more complete understanding of the factors that dictate inhibitor selectivity across the protein kinase family. We have used differential scanning fluorimetry to characterize inhibitor binding to the CDK active site. The profile of ΔTm values (the difference between the apparent melting temperature of the protein in the presence of the ligand and that determined for the protein alone) measured for a sufficiently diverse range of inhibitors is characteristic of the protein’s inhibitor binding properties and therefore represents a “fingerprint” of its active site. We have used such fingerprints to assess the chemical similarity of a number of CDK subfamily members, namely, CDK2, CDK4, CDK7, and CDK9. We have also used the technique to explore the inhibitor binding profiles of CDKs in different activation states to assess the extent to which CDK activation affects inhibitor binding.
Highlights
- A chemically diverse panel of kinase inhibitors was used to assess the chemical similarity of the ATP-binding sites of CDK subfamily members in a range of activation states
- Different activation states of a particular CDK may differ from each other as much as different CDKs in the same activation state
- Inhibitors discriminate more effectively among CDK family members in their monomeric state than in their cyclin-bound state
Comparison of the inhibitor-binding profiles of various cyclin-dependent kinase (CDK) subfamily members in different states of activation. Each block represents a CDK in a particular activation state. The internal consistency of inhibitory fingerprints for each CDK/state pair is indicated by a value within the corresponding block. The correlation of fingerprints between different CDK/state pairs is indicated by a value on the line that connects them.