CDK regulation
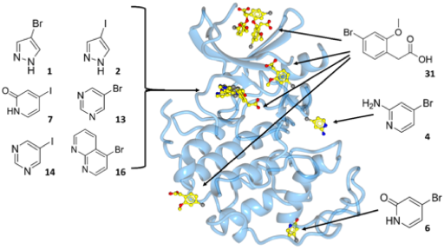
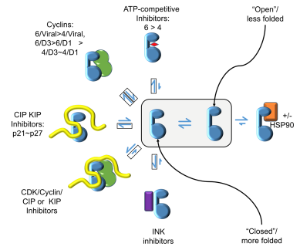
CDKs are positively and negatively regulated by multiple mechanisms. As examples of how structures can provide insights into how CDKs are regulated by reversible phosphorylation, we have determined structures for monomeric phosphorylated CDK2, CDK2-cyclin A inhibited by active site phosphorylation and CDK2-cyclin A bound to a peptide substrate. CDKs are also tightly regulated by reversible protein association. Characterisation of the cyclin recruitment (RXL) site has illustrated how CDK substrates and regulators compete for binding to the CDK-cyclin module. This competition offers the opportunity to integrate the signals from diverse signalling pathways to coordinate CDK activity with other processes in the cell. We have recently described an efficient and effective experimental approach to map potential protein interaction sites using a set of halogenated compounds expressing paired hydrogen-bonding motifs, termed FragLites. The power of the technique is illustrated by our results taking CDK2 as the target, and we are now using it to characterise other CDK-cyclin complexes. Finally, assembly of CDKs into active complexes is a tightly regulated by the Hsp90-Cdc37 chaperone system. We have demonstrated that CDK4 and CDK6 are differentially regulated by this system and hypothesize that there are multiple mechanisms whereby it may control formation of CDK4- and CDK6-cyclin complexes to impact their activities in normal vs transformed cell lines.