News
New Paper – Screening the COVID-19 main protease
Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease
https://www.biorxiv.org/content/10.1101/2020.05.27.118117v1
https://doi.org/10.1101/2020.05.27.118117
Alice Douangamath, Daren Fearon, Paul Gehrtz, Tobias Krojer, Petra Lukacik, C. David Owen, Efrat Resnick, Claire Strain-Damerell, Anthony Aimon, Péter Ábrányi-Balogh, José Brandaõ-Neto, Anna Carbery, Gemma Davison, Alexandre Dias, Thomas D Downes, Louise Dunnett, Michael Fairhead, James D. Firth, S. Paul Jones, Aaron Keely, György M. Keserü, Hanna F Klein, Mathew P. Martin, Martin E. M. Noble, Peter O’Brien, Ailsa Powell, Rambabu Reddi, Rachael Skyner, Matthew Snee, Michael J. Waring, Conor Wild, Nir London, Frank von Delft and Martin A. Walsh.
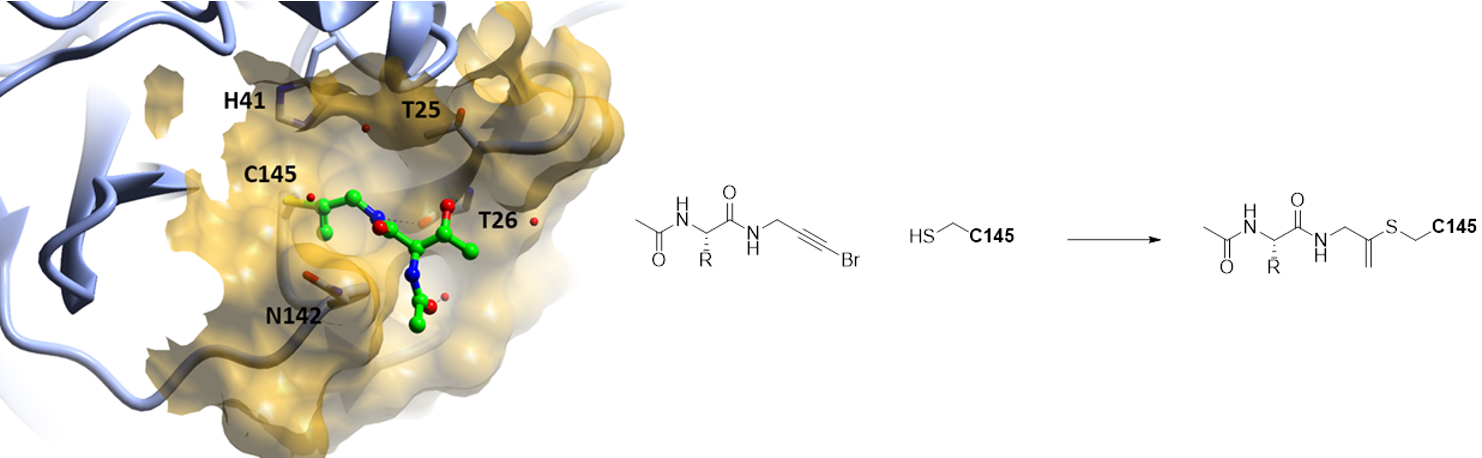
COVID-19, caused by SARS-CoV-2, lacks effective therapeutics. Additionally, no antiviral drugs or vaccines have been developed against the closely related coronavirus, SARS-CoV-1 or MERS-CoV.
The Newcastle Drug Discovery Group (Endicott, Noble and Waring) provided Diamond Light Source with new compound libraries synthesised in Newcastle’s medicinal chemistry laboratories to help identify starting points for such therapeutics. A large-scale screen of electrophile and non-covalent fragments was conducted through a combined mass spectrometry and X-ray approach against the SARS-CoV-2 main protease, one of two cysteine viral proteases essential for viral replication. The crystallographic screen identified 71 hits spanning the entire active site, as well as 3 hits at the dimer interface. Included in these hits were compounds synthesised by Newcastle PhD student Gemma Davison. These structures reveal routes to rapidly develop more potent inhibitors through merging of covalent and non-covalent fragment hits. Combined hits offer structural and reactivity information for on-going structure-based drug design against SARS-CoV-2 main protease.
Last modified: Mon, 19 Dec 2022 17:27:35 GMT