News
New Paper – Building a DNA-Encoded Library through Sequential Amide Coupling
RSC Med. Chem., 2025, 16, 4774-4780
DOI: 10.1039/D5MD00350D
https://pubs.rsc.org/en/content/articlelanding/2025/md/d5md00350d
Cameron E. Taylor, Grace Roper, Rhianna Young, Fredrik Svensson, Andreas Brunschweiger, Sam Butterworth, Andrew G. Leach and Michael J. Waring*
DNA-encoded libraries (DELs) are established as a highly effective and cost-efficient way to screen vast numbers of compounds for protein ligands, making them a key technology in modern drug discovery. By tagging molecules with DNA barcodes, DELs enable pooled screening and PCR-based detection, allowing large libraries to be built and tested rapidly with minimal protein use.
Despite their advantages, DEL synthesis is still carried out in relatively few labs as building very large libraries requires specialised equipment, logistics and expensive reagents thus creating barriers for smaller companies and academic groups. Moreover, extremely large libraries (>100 million members) often add little extra chemical diversity due to common scaffolds and can suffer from reduced fidelity and sequencing challenges. Smaller, structurally diverse DELs built with simple chemistry can overcome these limitations. By carefully selecting building blocks and avoiding excessive reliance on shared scaffolds, medium-sized libraries can achieve high chemical diversity while remaining more accessible and easier to validate.
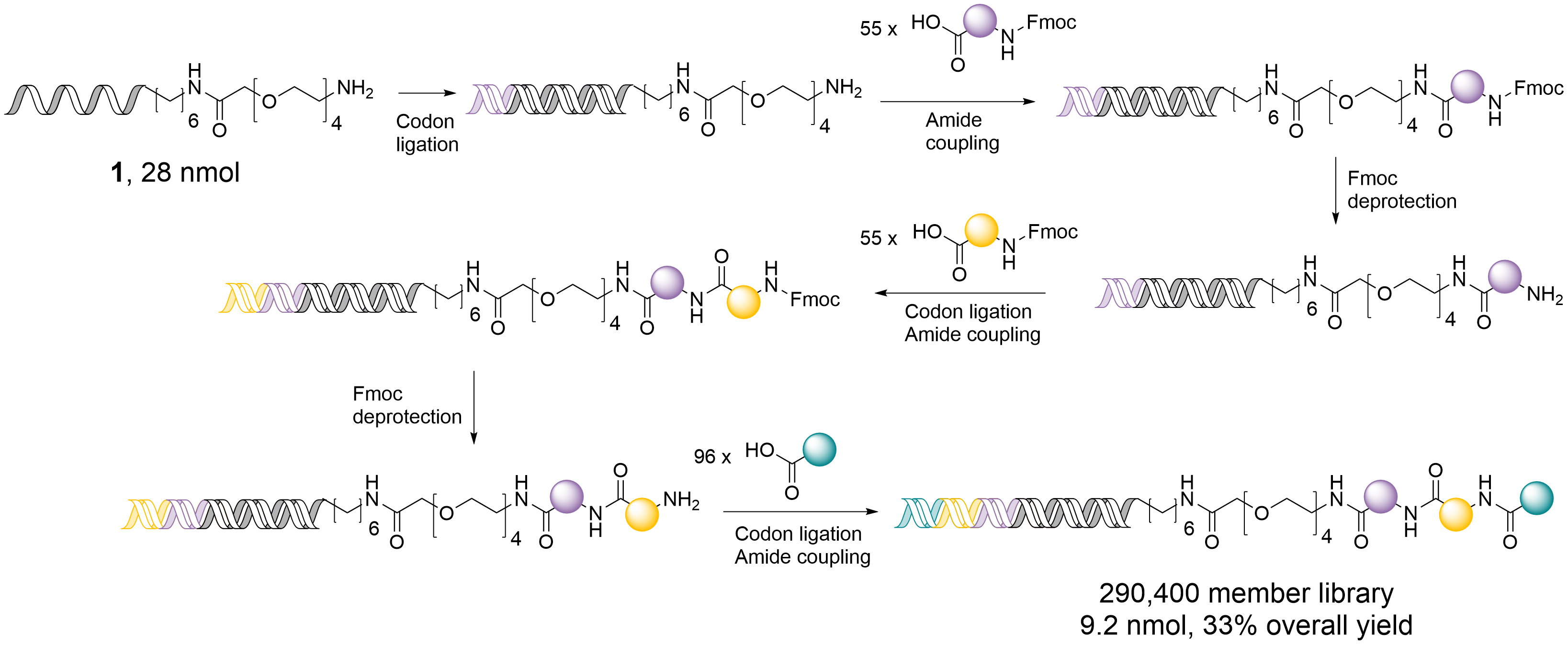
This work, led by Mike Waring and Cameron Taylor, set out to build two large DNA-encoded libraries (DELs) that capture lead-like chemical space relevant to early drug discovery. Using a straightforward, DNA-compatible amide coupling approach, the aim was to create libraries both chemically diverse and practical for broad screening applications. A split-and-pool DNA encoding strategy generated two libraries: one with around 300,000 compounds and an expanded version containing ~3 million. The libraries were constructed using sequential amide bond formation, chosen for its robustness and compatibility with DNA tags under aqueous conditions. Building blocks were carefully selected to ensure that the resulting compounds had lead-like physicochemical properties, including molecular weight, polarity and lipophilicity.
To verify the quality and functionality of the libraries, a screen against carbonic anhydrase IX (CA IX), a well-known exemplar protein, was performed. DNA sequencing allowed identification of enriched binders and confirmed the accuracy of the encoding process.
This work demonstrated that simple, reliable sequential amide coupling can be scaled efficiently to generate very large, chemically rich DNA-encoded libraries while preserving DNA integrity throughout synthesis. The resulting libraries showed broad chemical diversity that complements traditional high-throughput screening collections and provides access to novel regions of chemical space. Screening against carbonic anhydrase IX produced selective hits, confirming their value as starting points for medicinal chemistry. The approach relies on accessible building blocks and robust chemistry, lowering the barrier for other groups to create and apply DNA-encoded libraries in drug discovery.
Last modified: Tue, 23 Dec 2025 17:29:55 GMT