Using FragLites to map potential PPI
In contrast to protein active sites, which are refined by evolution to bind to other macromolecules, cofactors, or substrates and can generally be recognized from their shape and physicochemical properties, allosteric sites or sites of regulatory protein−protein interaction are harder to identify. Current small molecule libraries are designed to provide starting points for drug discovery and are not optimised to probe protein interaction sites by mimicking the groups that a protein would exploit to bind to a regulatory partner. In collaboration with Mike Waring and other chemistry colleagues, Martin Noble has designed a set of halogenated compounds expressing paired hydrogen-bonding motifs, termed FragLites. The FragLites identify protein interaction surfaces sensitively and unambiguously by X-ray crystallography, exploiting the anomalous scattering of the halogen substituent. The approach is illustrated by the FragLite map of the surface of CDK2, which includes the known orthosteric and allosteric sites and also four newly identified potential allosteric sites. We are now applying this approach to map the surfaces of other members of the CDK and cyclin families as a starting point to identify novel sites of protein interaction.
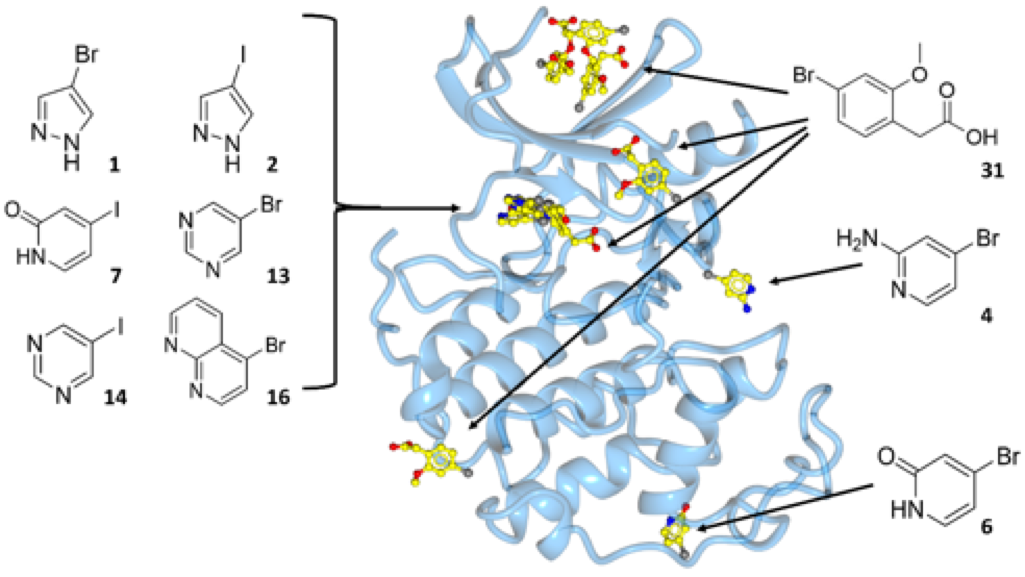
Binding sites of CDK2 revealed by FragLites. Overview of binding events to CDK2 (blue ribbon), depicting the six distinct sites identified by the FragLite library (yellow).
Collaborators: Mike Waring and colleagues at the NICR
Associated publication:
FragLites: minimal, halogenated fragments displaying pharmacophore doublets. An efficient approach to druggability assessment and hit (2019) Wood et al., J Med Chem 62: 3741-3752


