
NanoString Technology
The NanoString nCounter analysis system is the first and only platform that can deliver direct profiling of individual molecules in a highly multiplexed single reaction without any need for amplification. Critically, NanoString technology is superior to any other current RNA analysis technique for FFPE-derived material.
This protocol provides digital readouts of relative gene expression from hundreds of cDNA, mRNA, and miRNA fragments from extracted nucleic acids, FFPE material, and whole cell lysate, whilst obviating any enzymatic reactions that may introduce bias in the results.
Benefits of using the nCounter Platform
- Ability to measure 20 to 800+ genes in a single reaction
- Digitally detect and analyse samples in a fully automated manner
- No signal amplification or polymerase steps required
- High flexibility of sample input (including FFPE samples)
NanoString nCounter technology utilises an alternative approach to fluorescence-based expression compared to the intensity-based methods of Affymetrix and Illumina, as well as traditional methods such as Real Time Polymerase Chain Reactions (RT-PCR). Rather than calculating an intensity score to determine relative expression levels, NanoString directly profiles individual molecules in a highly multiplexed reaction by assigning fluorescently labeled probes to genes of interest. In this manner, probes that have bound to a molecule of interest can be counted by a computerised optical lens. A chain of 7 fluorescent tags are bound to a biotin-containing oligo complementary to a 100bp region of the 3’ end of a gene of interest. The order of these coloured tags act as a ‘barcode’ to indicate which gene the complementary oligo refers to.
Unlike other gene expression methods, NanoString does not require any pre-amplification or polymerisation of sample input material. As a result, issues with amplification bias (where even small deviations in molecule amplification in a heterogenous mix are amplified over a number of cycles, resulting in comparative over- or under-amplification of certain sequences and therefore a skewing of any true underlying expression differences) are non-existent. The random nature of probe binding to the detection surface of the nCounter cartridges eliminates region specific gene drop-outs, while the non-reliance on fluorophore signal intensity negates compensation based errors. Fully automated loading of the cartridge, pre-processing, digital detection and analysis minimizes user error, making NanoString assays reproducible, highly sensitive and extremely robust.
While fluorescence-based, NanoString expression technology uses a chain of fluorescent tags as ‘barcodes’ for a specific mRNA transcript of interest. Over 800 genes can be assessed per sample in this manner for up to 12 samples per cartridge. As NanoString Technology does not rely on signal intensity or fluorescence scores, there is very little bias introduced. Instead, NanoString uses biotin molecules to bind captured mRNA hybridised-reporter probe strands to the streptavadin-coated surface of the cartridge. Once linearised by induction of an electrical gradient, the ‘barcodes’ can be optically detected by a computerised camera and microscope. From this, individual mRNA strands can be detected and identified as representing a specific gene. Each captured probe is counted to give the actual number of mRNA strands that were captured in each sample. This highly specific direct profiling and digital detection process produces large quantities of robust data in a short period of time.
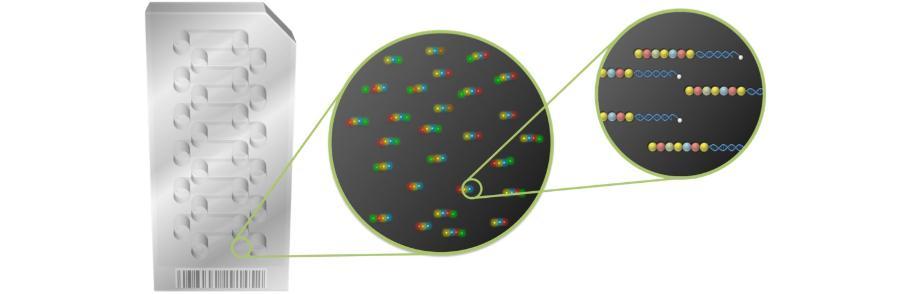
Schematic representation of NanoString nCounter expression array
For more information about the technology, please visit: www.nanostring.com/applications/technology
