Experiments we perform
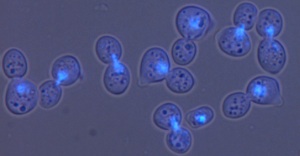
In the image above on the right a culture of cdc13-1 mutants has been incubated at 36oC for 2 hours, the DNA stained with a blue dye and the cells photographed using a microscope. Most cells show two buds and a single bright blue DNA mass because in these cells the telomeres are uncapped and segregation of DNA in mitosis is stopped by DNA damage checkpoint pathways.
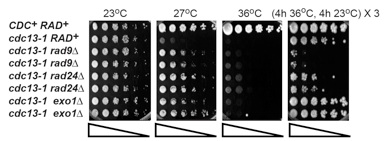
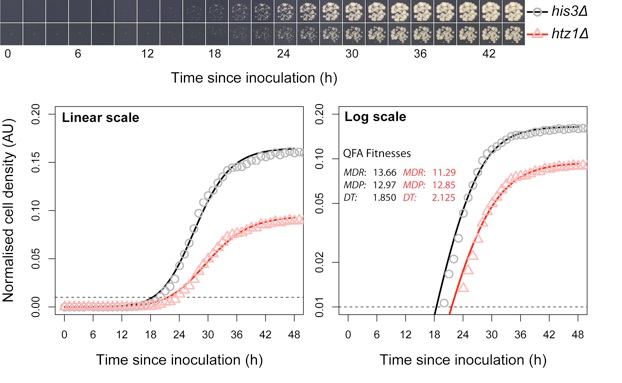
Use of automated camera system and image analysis allows us to convert spot tests (as above) into growth curve. We can perform thousands of such measurements in parallel, in Quantitative Fitness Analysis.
We use many other methods too. Several years ago we developed QAOS (Quantitative Amplification of Single Stranded DNA), a real time PCR based method to measure single stranded DNA arising in yeast cells. QAOS and many of the other experimental approaches we use are described in our publications.