| Molecular and Cellular Evolution of Microbial Eukaryotes |

| Introduction |
| People |
| Publications |
| Downloads |
| Newcastle |
| EMBO World 09 |
| Molecular Evolution Workshop 2010 |
| Trachipleistophora hominis |
Dr. Naiara Rodriguez-Ezpeleta |  |
Contact information
- Phone number:
- e-mail adress: nrodriguez (at) cicbiogune.es
Work description
Background
After graduating in Biology from the University of Basque Country, I moved to Montreal (Quebec, Canada) to work in Franz Lang's group. During my PhD, I focused on the study of the relationships among eukaryotic groups using genome-scale phylogenetics.
For this goal, I spent the first part of my PhD developing an efficient protocol to construct cDNA libraries from glaucophytes, jakobids and malawimonads, three evolutionarily important but poorly studied eukaryotic groups.
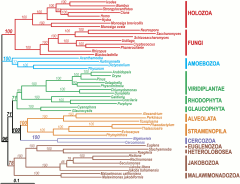
In the second stage of my PhD, I focussed on using phylogenomics to address fundamental questions in eukaryotic evolution such as the origin of choroplasts and the phylogenetic position of the enigmatic jakobids and malawimonads.
I also performed studies on the impact of tree reconstruction artefacts on genome-scale phylogenetic analyses. Using two empirical examples, I showed situations where phylogenomic results were contradictory and explored different methods to detect and overcome the systematic errors that were responsible of the misleading results.
My PhD thesis can be downloaded here.
Current work
During my time as a post doc, I will focus on two fundamental questions of eukaryotic evolution: the origin of mitochondria and the position of the eukaryotic root.
Teaching
During the summers of 2005 and 2006 I was a Teaching Assistant for the Workshop on Molecular Evolution in Woods Hole, USA. I have also participated as Teaching Assistant for graduate courses at the Université de Montréal and for the Bioneq Phylogenomics Workshop organized at the Université du Québec á Montréal.
Naiara is now working in the Genome Analysis Group in CIC bioGUNE back home in the Basque country, her new contact details are:
Dr Naiara Rodriguez-Ezpeleta
Functional Genomics Unit
CIC bioGUNE
Parque Tecnologico de Bizkaia, Ed. 502
48160 Derio, Bizkaia
Spain
Naiara's publications
- Rodriguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H and Lang BF. Toward the resolution of the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. vol 17 p. 1420-5 (2007). pubmed
- Rodriguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang BF and Philippe H. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst Biol. vol 56 p. 389-99 (2007). pubmed
- Rodriguez-Ezpeleta N, Philippe H, Brinkmann H, Becker B and Melkonian M. Phylogenetic analyses of nuclear, mitochondrial, and plastid multigene data sets support the placement of Mesostigma in the Streptophyta. Mol Biol Evol. vol 24 p. 723-31 (2007). pubmed
- Roure B, Rodriguez-Ezpeleta N and Philippe H. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. vol 7 (2007). pubmed
- Rodriguez-Ezpeleta N and Philippe H. Plastid origin: replaying the tape. Curr Biol. vol 16 p. R53-6 (2006). pubmed
- Rodriguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, Bohnert HJ, Philippe H and Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. vol 15 p. 1325-30 (2005). pubmed
- Leigh J, Seif E, Rodriguez-Ezpeleta N, Jacob Y, Lang BF. Fungal evolution meets fungal genomics. In Handbook of Fungal Biotechnology, 2nd ed. (D. Arora, ed.) pp. 145-161. Marcel Dekker Inc. New York, NY. (2003).