| Molecular and Cellular Evolution of Microbial Eukaryotes |

| Introduction |
| People |
| Publications |
| Downloads |
| Newcastle |
| EMBO World 09 |
| Molecular Evolution Workshop 2010 |
| Trachipleistophora hominis |
Dr. Cedric Simillion |  |
Contact information
- Phone number:
- e-mail adress: cedric.simillion (at) psb.vib-ugent.be
Work description
Background
I have an MsC. in biotechnology from Ghent University and did my PhD the bioinformatics lab of Yves Van de Peer in Ghent, Belgium. During my PhD, I have mainly worked on the structural evolution of genomes. My research involved developing software (i-ADHoRe) to detect traces of ancient large-scale gene duplication events in various genomes such as the flowering plants Arabidopsis thaliana, Oryza sativa and bony fish Takifugu rubripes
Current work
|
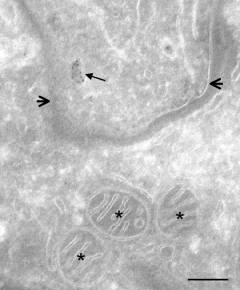
| The remnant mitochondrion (mitosome) of Trachipleistophora hominis. The short arrows define the border of the microsporidian, which is living inside a rabbit kidney cell, and the stars indicate the rabbit kidney cell mitochondria. The long arrow indicates the Trachipleistophora mitosome which reacts strongly with the specific antibody to the Trachipleistophora mitochondrial Hsp70. From Embley et al. 2003. |
The goal of my research is to investigate the structure and function of these mitochondrial homologues by comparing the mitochondrial proteomes and interactomes of a broad set of eukaryotic proteomes, including both organisms with bona fide mitochondria as well as organisms featuring hydrogenosomes and mitosomes.
Apart from the people in our group, I also collaborate with Jennifer Hallinan and Neil Wipat for the analysis of interactomes.
Cedric has just moved back to Belgium to work in the Department of Plant Systems Biology, Ghent University, his new contact details are:
Dr Cedric Simillion
VIB Department of Plant Systems Biology
Ghent University
Technologiepark 927
9052 Ghent
Belgium
Cedric's publications
- Simillion C, Janssens K, Sterck L and Van de Peer Y. i-ADHoRe 2.0: an improved tool to detect degenerated genomic homology using genomic profiles. Bioinformatics. vol 24 p. 127-8 (2008). pubmed
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Muller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM and Johnson PJ. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. vol 315 p. 207-12 (2007). pubmed
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S and Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. vol 7 p. R43 (2006). pubmed
- Gevers D, Vandepoele K, Simillion C and Van de Peer Y. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. vol 12 p. 148-54 (2004). pubmed
- Simillion C, Vandepoele K and Van de Peer Y. Recent developments in computational approaches for uncovering genomic homology. Bioessays. vol 26 p. 1225-35 (2004). pubmed
- Simillion C, Vandepoele K, Saeys Y and Van de Peer Y. Building genomic profiles for uncovering segmental homology in the twilight zone. Genome Res. vol 14 p. 1095-106 (2004). pubmed
- Vandepoele K, Simillion C and and Van de Peer Y. The quest for genomic homology. Curr. Genomics. vol 5 p. 299-308 (2004).
- Raes J, Vandepoele K, Simillion C, Saeys Y and Van de Peer Y. Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct Genomics. vol 3 p. 117-29 (2003). pubmed
- Vandepoele K, Simillion C and Van de Peer Y. Evidence that rice and other cereals are ancient aneuploids. Plant Cell. vol 15 p. 2192-202 (2003). pubmed
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M and Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci U S A. vol 99 p. 13627-32 (2002). pubmed
- Vandepoele K, Saeys Y, Simillion C, Raes J and Van De Peer Y. The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. vol 12 p. 1792-801 (2002). pubmed
- Vandepoele K, Simillion C and Van de Peer Y. Detecting the undetectable: uncovering duplicated segments in Arabidopsis by comparison with rice. Trends Genet. vol 18 p. 606-8 (2002). pubmed