News & Events
New Preprint
New Preprint: 'Evolutionary Constraint Genes Implicated in Autism Spectrum Disorder Across 2,054 Nonhuman Primate Genomes' Now Available on bioRxiv
Background:
Significant progress has been made in elucidating the genetic underpinning of Autism Spectrum Disorder (ASD). This childhood-onset chronic disorder of cognition, communication and behaviour ranks among the most severe from a public health perspective, and it is therefore hoped that new discoveries will lead to better therapeutic options. However, there are still significant gaps in our understanding of the link between genomics, neurobiology and clinical phenotype in scientific discovery. New models are therefore needed to address these gaps. Rhesus macaques (Macaca mulatta) have been extensively used for preclinical neurobiological research because of remarkable similarities to humans across biology and behaviour that cannot be captured by other experimental animals.
Methods:
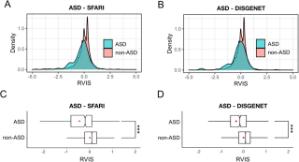
We used the macaque Genotype and Phenotype (mGAP) resource (v2.0) consisting of 2,054 macaque genomes to examine patterns of evolutionary constraint in known human neurodevelopmental genes. Residual variation intolerance scores (RVIS) were calculated for all annotated autosomal genes (N = 18,168) and Gene Set Enrichment Analysis (GSEA) was used to examine patterns of constraint across ASD genes and related neurodevelopmental genes.
Results:
We demonstrated that patterns of constraint across autosomal genes are correlated in humans and macaques, and that ASD-implicated genes exhibit significant constraint in macaques (p = 9.4 x 10-27). Among macaques, many key ASD genes were observed to harbour predicted damaging mutations. A small number of key ASD genes that are highly intolerant to mutation in humans, however, showed no evidence of similar intolerance in macaques (CACNA1D, CNTNAP2, MBD5, AUTS2 and NRXN1). Constraint was also observed across genes implicated in intellectual disability (p = 1.1 x 10-46), epilepsy (p = 2.1 x 10-33) and schizophrenia (p = 4.2 x 10-45), and for an overlapping neurodevelopmental gene set (p = 4.0 x 10-10)
Limitations:
The lack of behavioural phenotypes among the macaques whose genotypes were studied means that we are unable to further investigate whether genetic variants have similar phenotypic consequences among nonhuman primates.
Conclusion:
The presence of pathological mutations in ASD genes among macaques, and the evidence of similar constraints in these genes to humans, provide a strong rationale for further investigation of genotype-phenotype relationships in nonhuman primates. This highlights the importance of identifying phenotypic behaviours associated with clinical symptoms, elucidating the neurobiological underpinnings of ASD, and developing primate models for translational research to advance approaches for precision medicine and therapeutic interventions.
Last modified: Wed, 22 Nov 2023 19:25:58 GMT






