| Molecular and Cellular Evolution of Microbial Eukaryotes |

| Introduction |
| People |
| Publications |
| Downloads |
| Newcastle |
| EMBO World 09 |
| Molecular Evolution Workshop 2010 |
| Trachipleistophora hominis |
Introduction
About us
The research group is led jointly by Martin Embley and Robert Hirt. Members of the group are interested in the evolution of eukaryotic cells, their genomes and organelles, namely hydrogenosomes and mitosomes. We have focused on anaerobic and parasitic eukaryotes including Entamoeba, Giardia, Trichomonas, and microsporidia.
Members of the group have a range of skills ranging from cell biology, molecular biology, phylogenetics and computational biology. We have a good mix of nationalities in the group, which makes for a friendly, supportive and multicultural work environment.
News
Open positions
We often have research positions and information about these can be obtained by contacting Martin or Robert. We also have an excellent track record in obtaining EU funding including the generous International (non-European only) and Intra-European (European only) Marie Curie Fellowships, so if our research interests you and you would like to apply for one of these, then please get in touch for details.
Research
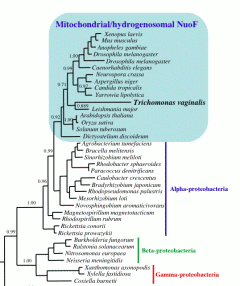
|
| Phylogenetic tree showing that, after recoding data, the Trichomonas hydrogenosomal NuoF protein clusters with bona-fide mitochondrial proteins. From Hrdy et al. 2004. |
Genomics and Bioinformatics
We are using whole genome data to investigate the contrasting roles of vertical and horizontal inheritance, including identifying genes of endosymbiotic origin, in shaping eukaryotic genomes. We are collaborating Neil Hall (University of Liverpool) to sequence to genomes of parasitic eukaryotes. We have a long term collaboration with Thomas Sicheritz-Ponten (CBS, Denmark) to develop automated genome-wide analyses for detecting horizontal gene transfer. With Yves Van de Peer's group (University of Gent, Belgium) we are investigating the core mitochondrial proteome. We also collaborate with Jim Warwicker (University of Manchester) on protein structure modelling of specific proteins or domains. An ongoing collaboration with Neil Wipat (School of Computing Science, Newcastle) aims to use comparative genomics to investigate the protein repertoire underlying the pathogenicity of mucosal microbes. We collaborate with Jane Carlton (New York University, Langone Medical Center, USA) to investigate the unexpectedly large and repetitive genome of Trichomonas vaginalis.
Phylogenetics
We are developing better methods, based upon likelihood and Bayesian approaches, for phylogenetic analysis of molecular data. Our aim is to improve the reconstruction of phylogenetic trees relevant to understanding the early evolution of eukaryotes and the origins of eukaryotic genes. We collaborate with Peter Foster (The Natural History Museum, London) and Tom Nye (Newcastle University) on these projects.
|
| Location of iron-sulphur cluster assembly proteins in the mitosomes of Encephalitozoon cuniculi. For full details see Goldberg et al. 2008. Nature 452: 624-628 |
Cell Biology and Cellular Parasitology
We are investigating the functions of organelles - hydrogenosomes and mitosomes - related to mitochondria. Our investigations are aimed at identifying the essential functions of these organelles for eukaryotes and are guided by our bioinformatics studies.
We collaborate with Jan Tachezy and his group in Prague, Edmund Kunji (Cambridge University), Roland Lill (Philipps-University, Marburg) and Adrian Hehl (University of Zurich) in this area of work. We also collaborate with John Lucocq on the ultrastructure of mitochondria derived organelles. To try and understand the pathogenic mechanisms of Trichomonas, in particular how it colonizes its host, we collaborate with Pier-Luigi Fiori (University of Sassari, Italy) and Hugo Lujan (School of Medicine, Catholic University of Cordoba, Argentina).
Where are we?

|
| The Institute for Cell and Molecular Biosciences (ICAMB) where our team is located. |
Newcastle is a small but vibrant city with a large student population, lots of bars and restaurants, and cheap accommodation. It is situated in the North of England close to wonderful countryside full of history, the Roman Wall is close by, and to beautiful but largely deserted beaches (this is after all England). Edinburgh in Scotland is only a short rail journey away and London can be reached in less than three hours by train. Newcastle has an international airport with good links to continental Europe.
The research group is located in the Institute of Cell and Molecular Biosciences, a 5* research centre in the Faculty of Medical Sciences. The Faculty provides excellent infrastructure, including state of the art computing and imaging facilities for our work. The University has an excellent library and a full range of e-journals.
Teaching Materials
We teach an international course on Molecular Systematics with Mark Wilkinson and Peter Foster (The Natural History Museum) and James O McInerney (University of Maynouth, Ireland). The course has been running since 1996 in different countries including Brazil, Switzerland and Finland. Please get in touch if you and your institute are interested in hosting the course in your own country. In the past, the course has been funded by: The British Council, The Wellcome Trust and EMBO. The course material can be freely downloaded for teaching or research from the page Molecular Evolution Workshop Brazil 2010.
